InTAD: chromosome conformation guided analysis of enhancer target genes
Konstantin Okonechnikov, Serap Erkek, Jan O. Korbel, Stefan M. Pfister & Lukas Chavez
A fundamental concept of chromatin organization is topologically associated domains (TADs). TADs are segments of the genome characterized by frequent chromosome interactions within themselves and they are insulated from adjacent TADs
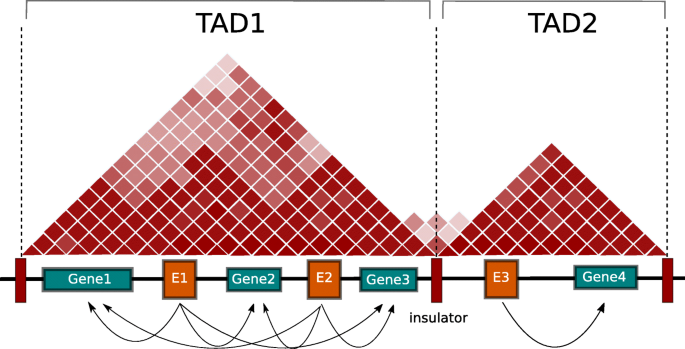
Chromatin is organized in topologically associated domains (TADs). The InTAD software package tests for significant correlations between genes and enhancers restricted by TAD boundaries
chromatin is organized in topologically associated domains (TADs). While TADs are relatively stable across cell types, intra-TAD activities are cell type specific. Epigenetic profiling of different tissues and cell-types has identified a large number of non-coding epigenetic regulatory elements (‘enhancers’) that can be located far away from coding genes. Linear proximity is a commonly chosen criterion for associating enhancers with their potential target genes. While enhancers frequently regulate the closest gene, unambiguous identification of enhancer regulated genes remains to be a challenge in the absence of sample matched chromosome conformation data.

